
| Research History |
| Software Structure |
| Specie Sensitivity Distribution |
| BAYESIAN Inference |
| MCMC Simulation |
| DIC Optimization |
| Ecorisk & Uncertainty |
| Joint Probability Curve |
| Exergy SSD |
| Main Function Lists Panel |
| BMC-SSD Panel |
| Models Optimization Panel |
| JPC Panel |
| ExSSD Panel |
| Work Path & Output Results |
| Installation & Initialization |
| Folder & File Extraction |
| SSD Models & Ecorisk |
| JPC & Its Indicators |
| Models Optimization & Parameters |
| ExSSD Models & ExEcorisk |
Links
| College of Urban and Environment Science |
| Peking University |
Exergy SSD
The damage of the ecosystem can exhibit by the relative changes of Exergy (ΔEx), which is shown in formula (3). Where the biomass loss of each species i can be represented by dose-effect curve F(c) (Formula (4)). The resistance of each species to the toxicity of chemicals is usually expressed by L(E)C50, and when c equals to L(E)C50, 50% of organisms will loss the bioactivities, and F(c) = 0.5. However, the biomass of each species is hard to get in the ecosystem, therefore, Wang (2009) proposed to divide the organisms into algae, invertebrates, and vertebrates, corresponding to there trophic levels. A specific trophic level includes different species j. It is assumed that their biomass is similar, and the Bi,mean is their mean value, and B1,i is calculated by formula (5), where j1 of species are not affected, and the affected ratio of j2 of species are related to L(E)C50. The toxicity data of these species are easily to obtain. The potential affected ratio of species is PAFi, and the exposure concentration of each species is c, then ΔEx can be represented by formula (6), where the coefficient “1/2” is from the half-lethal dose. Due to the difficulty of obtaining the absolute biomass, the relative biomass bi is substituted, and ki value is normalized according to Wang’s method (2009). Therefore, this software uses the relative exergy variation (δEx) to represent for the ecological risks at system level (formula (7)).
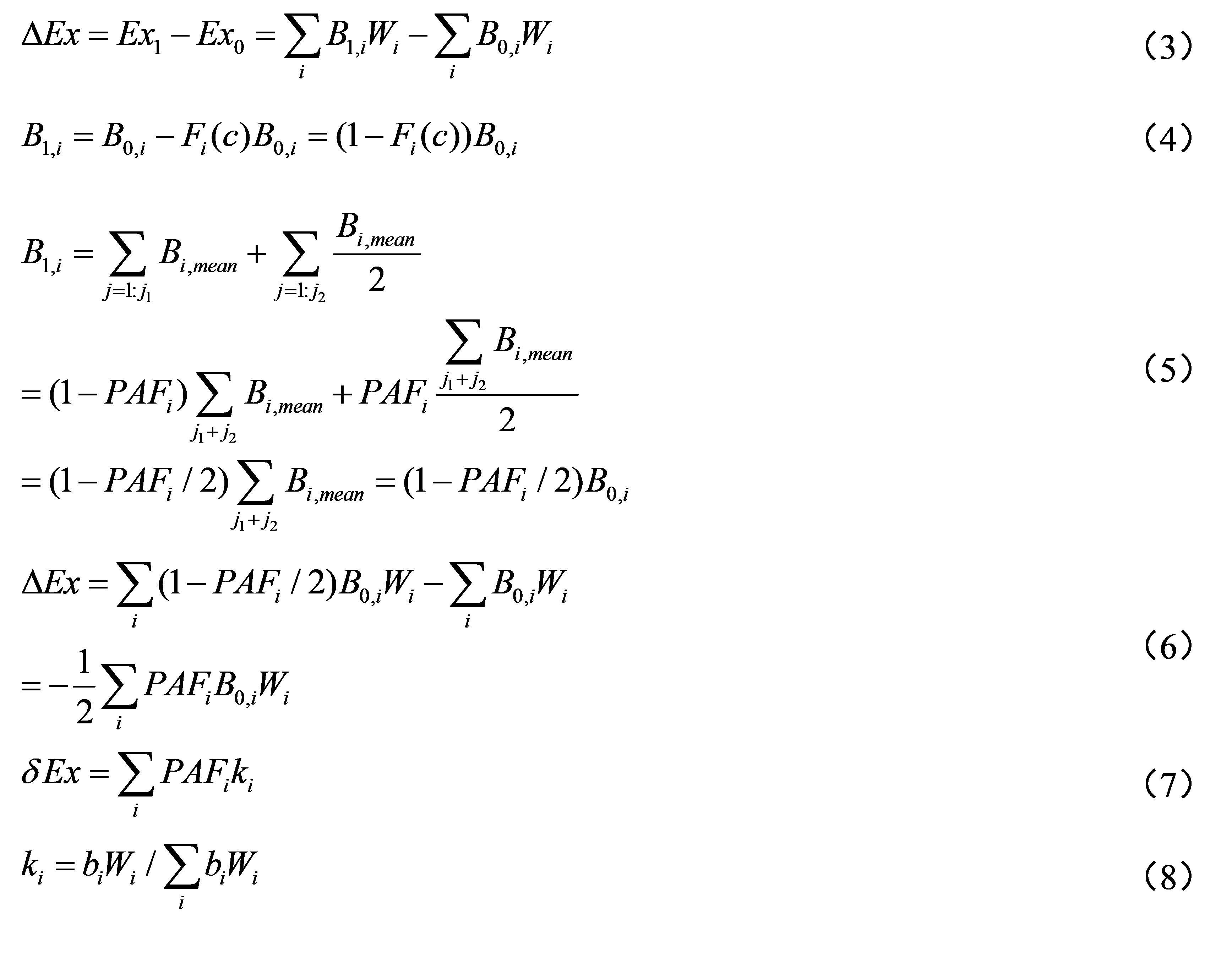
Above noted formulas, B0,i is the absolute biomass of species group i, and Wi is the exergy weight of chemical group i, which is the β value in the structural dynamics model. To establish the SSD at system level in this software, the determination steps of k value are:
(1) to establish the distributions of the three trophic levels, according to their Wi and bi values collected by He (2014), and determine the best distribution by BMC-SSD;
(2) to random sampling N times (10000 by default) by using Monte Carlo simulation from above distributions, and calculate the k value using formula (8) to get the it distribution and related statistics;
(3) to calculate the uncertainty boundaries of ExSSD or δEx value and related statistics at high, median, and low level using the SSD or PAF values determined by BMC-SSD at three trophic levels, k value distribution (in step 2), and formula (6).
![]()